There are a lot of ways to make R packages. Many blog posts have covered making R packages, but for the most part they’ve covered only how they make packages, going from the required files for a package, what to put in DESCRIPTION, etc. But what about the tooling? I’m not going to talk about the code, etc. - but rather the different ways to approach it.
The blog posts/etc. on making R packages:
- Writing an R package from scratch - Hilary Parker
- Developing R packages - Jeff Leek
- stat545 - Write your own R package - Jenny Bryan’s statistics 545 class at UBC
- R package primer - Karl Boman
- Making Your First R Package - Fong Chun Chan
- R Package Development Pictorial - Matthew Denny
- Coursera course on building R packages
- R Packages by Hadley for a full treatment of the subject.
- From time to time you may need to reference CRAN’s Writing R Extensions.
the ways
The following are not mutually exclusive - some can be combined with others.
In process of writing this I figured I should ask other people what they do. I ended up asking 16 people - not a random selection or a big enough n to really say anything conclusively. But it did allow me to cover more ways of doing
package dev.
mason
github: https://github.com/metacran/mason (by Gábor Csárdi)
Note that
masonis on CRAN, but it’s a completely different package.
You can use mason inside of R or via Rscript on the cli.
devtools::install_github("metacran/mason")
library(mason)
mason::mason()
Then you’ll go through a series of prompts asking you for inormation (package name, license, your name, etc.)
Out of 16 people I talked to, 2 mentioned using mason.
devtools
github: https://github.com/hadley/devtools (by Hadley Wickham)
Within R:
install.packages("devtools")
library(devtools)
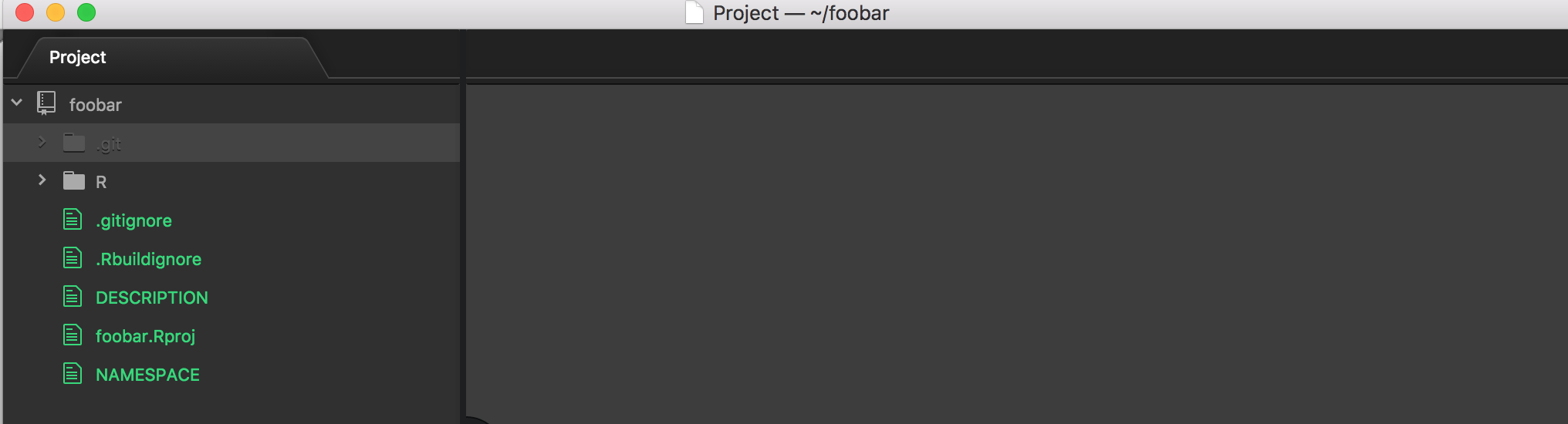
devtools::create("foobar")
On the cli, we can do:
Rscript -e 'devtools::create("foobar")'
devtools::create() adds basic set of files needed for an R package - and also adds files assuming you use the RStudio IDE. Though you can not add RStudio files by choosing rstudio = FALSE.
Be aware of the default entry in the NAMESPACE file: exportPattern("^[^\\.]"). The first time you generate documentation, e.g., via devtools::document() your NAMESPACE file will be changed to only export those things you want exported, which is ideal.
Out of 16 people I talked to, 7 mentioned using devtools.
rcmdcheck
rcmdcheck: https://github.com/r-lib/rcmdcheck
This is an alternative to running R CMD CHECK or devtools::check(), that gives nice colorized output, at least in the terminal.
I usually run it like this in the root of an R package directory in my terminal (running with --as-cran to check as CRAN does, and --run-dontrun to run examples wrapped in \dontrun{}):
Rscript -e 'rcmdcheck::rcmdcheck(args = c("--as-cran", "--run-dontrun"))'
IDE: RStudio
rstudio: https://www.rstudio.com/products/rstudio/#Desktop
The following is a guide provided by RStudio for creating packages in RStudio IDE: Using RStudio for package development
When in RStudio - New Project in upper left hand corner - choose new or existing directory - choose R package - name the package, and you probably want to initialize git by checking the appropriate box.
Out of 16 people I talked to, 14 used RStudio all the time or most of the time. It’s popular, to say the least.
A noteable quote from one person I talked to:
all rstudio all day
RStudio inside of Docker
rstudio server: https://www.rstudio.com/products/rstudio/download-server/ rstudio server docker container: https://hub.docker.com/r/rocker/rstudio/
I know of at least one person that works this way, and surely there are others.
As simple as:
docker run -d -p 8787:8787 rocker/rstudio
Then visit localhost:8787 in your browser.
web
For example, we could just create files from the Github website. e.g,.
- New Repository
- then add files you’d need for an R package and edit those in the browser
Out of 16 people I talked to, 2 mentioned starting with creating a GitHub repository, then pulling that down, R development, etc. etc., then push back up. But no one mentioned all in browser - although see phone dev below.
The Github web interface is an important starting point for getting people into code in general when they are not familiar with git.
text editor
If you primarily work in a text editor perhaps this (using Atom editor):
Rscript -e 'devtools::create("foobar")' && cd foobar && git init && atom .
Or the same for Sublime Text with subl . instead of atom .
Two of 16 people I talked to mentioned using Emacs exclusively or mostly. One of the 16 people mentione Sublime Text by name - that’s also the editor I use (I often have RStudio and Sublime Text open for the same R package - switching between them for the features I like).
copy/paste
Sometimes when creating a new package I know of a previous package I’ve created that may have similar code I want in the new one or so. So I just copy/paste essentially the old package into a new folder. Be careful when doing this: make sure to delete git history, then re-initialize git (rm -rf .git && git init in the new repository). Ideally you use roxygen/devtools for docs - in which case just delete all files in man/ then when you generate docs, you get all new man files.
rhub
rhub: https://builder.r-hub.io/ https://github.com/r-hub https://www.r-consortium.org/events/2016/10/11/r-hub-public-beta
R-hub is a project by Gabor Csárdi, funded by the R Consortium, which is a service for developing, building, testing and validating R packages.
One of the 16 people I talked to mentioned rhub - but I imagine many of them use it. I use it :)
I usually use it from the command line (or you can use it from within R, either on CLI or RStudio), like rhub::check_for_cran() to get checks for my package on Windows and two Linux platforms, before sending to CRAN.
the Makefile
The Makefile is a file containing a set of directives. Some use a Makefile for a few or even most things one does in package development, from re-making man files, to building, installing, checking, building vignettes, making pkgdown docs, and more. Makefiles can include actions that do not just R things, but run other programming/command line tools. It’s a good idea when contributing to another R package to look for a Makefile - and to use them in your own package development. I don’t personally use them enough, and ideally will use them more in the future.
Here’s an example of a Makefile from Rich FitzJohn:

ruby?
To scratch a personal itch, I made a little Ruby gem with a command line tool to run one or more specific tests by fuzzy matching the name of the test file. Reason is, sometimes I work on a test file and I just want to run that test and not any others - and not from within RStudio, but form the terminal.
https://github.com/sckott/rubrb
➜ rb test config
using:
tests/testthat/test-config-fxns.R
config fxns: ........
DONE ===========================================================================
I’m sure there’s lots of these types of things out there - scratching an itch that helps the person work the way they want to work.
While we’re on the topic of Ruby, Travis-CI has a nice Ruby gem travis to interact with Travis for your R packages. There’s also one for Circle-CI and I’ve written one for Appveyor.
phone dev
DO NOT TRY THIS AT HOME
tracestack: search Stack Overflow for your most recent error msg. First #rstats package written entirely on a phone? https://t.co/IRX2luiR0N
— David Robinson (@drob) April 25, 2015
.@drob just wrote an #rstats package from his phone, in case you wanted to feel even less productive checking twitter
— Hilary Parker (@hspter) April 25, 2015
So that idea for throwing traceback to stackoverflow? @drob is actually writing it. As a package. Live on github. ON HIS PHONE. #rstats
— Oliver Keyes (@kopshtik) April 25, 2015
putting stuff on the web
While we’re talking about tooling, I thought I should briefly mention putting code up on the interwebs. There’s many code hosting options - for brevity, we’ll just cover GitHub.
It’s a good idea to learn command line git, and related command line tools that make using git easier - if you can get work done faster you have more time to look at cat pictures!
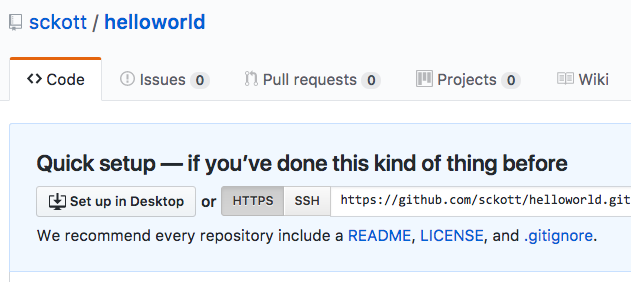
hub is one git tool that I use a lot. For example, create a folder, initialize a git repo, push to github, then open the just created repo on Github:
mkdir helloworld && cd helloworld && hub init && hub create sckott/helloworld && hub browse
where hub create uses the owner/repo pattern