I posted last week a simple function to plot networks using ggplot2 package. Here is version 2. I still need to work on figuring out efficient vertex placement.
Changes in version 2:
- You have one of three options: use an igraph object, a matrix, or a dataframe (matrices will be converted to data frames within the function)
- If you have data on food webs similar to that provided in the Takapoto dataset provided in the NetIndices package, you can set trophic = “TRUE”, and gggraph will use the function TrophInd to assign trophic levels (the y axis value) to each vertex/node. You have to provide additional information along with this option such as what the imports and exports are, see NetIndices documentation.
- I added some simple error checking.
- if using method=“df” and trophic=“FALSE”, x axis placement of vertices is now done using the function degreex (see inside the fxn), which sorts vertices according to their degree (so the least connected species are on the left of the graph; note that species with the same degree are not stacked on the y-axis because e.g., two vertices of degree=5 would get x=3 then x=4).
# ggraph Version 2
require(bipartite)
require(igraph)
require(ggplot2)
# gggraph, version 3 g = an igraph graph object, a matrix, or data frame
# vplace = type of vertex placement assignment, one of rnorm, runif, etc.
# method = one of 'df' for data frame, 'mat' for matrix or 'igraph' for an
# igraph graph object trophic = TRUE or FALSE for using Netindices
# function TrophInd to determine trophic level (y value in graph)
# trophinames = columns in matrix or dataframe to use for calculating
# trophic level import = named or refereced by col# columns of matrix or
# dataframe to use for import argument of TrophInd export = named or
# refereced by col# columns of matrix or dataframe to use for export
# argument of TrophInd dead = named or refereced by col# columns of matrix
# or dataframe to use for dead argument of TrophInd
gggraph <- function(g, vplace = rnorm, method, trophic = "FALSE", trophinames,
import, export) {
degreex <- function(x) {
degreecol <- apply(x, 2, function(y) length(y[y > 0]))
degreerow <- apply(x, 1, function(y) length(y[y > 0]))
degrees <- sort(c(degreecol, degreerow))
df <- data.frame(degrees, x = seq(1, length(degrees), 1))
df$value <- rownames(df)
df
}
# require igraph
if (!require(igraph))
stop("must first install 'igraph' package.")
# require ggplot2
if (!require(ggplot2))
stop("must first install 'ggplot2' package.")
if (method == "df") {
if (class(g) == "matrix") {
g <- as.data.frame(g)
}
if (class(g) != "data.frame")
stop("object must be of class 'data.frame.'")
if (trophic == "FALSE") {
# data preparation from adjacency matrix
temp <- data.frame(expand.grid(dimnames(g))[1:2], as.vector(as.matrix(g)))
temp <- temp[(temp[, 3] > 0) & !is.na(temp[, 3]), ]
temp <- temp[sort.list(temp[, 1]), ]
g_df <- data.frame(rows = temp[, 1], cols = temp[, 2], freqint = temp[,
3])
g_df$id <- 1:length(g_df[, 1])
g_df <- data.frame(id = g_df[, 4], rows = g_df[, 1], cols = g_df[,
2], freqint = g_df[, 3])
g_df_ <- melt(g_df, id = c(1, 4))
xy_s <- data.frame(degreex(g), y = rnorm(length(unique(g_df_$value))))
g_df_2 <- merge(g_df_, xy_s, by = "value")
} else if (trophic == "TRUE") {
# require NetIndices
if (!require(NetIndices))
stop("must first install 'NetIndices' package.")
# data preparation from adjacency matrix
temp <- data.frame(expand.grid(dimnames(g[-trophinames, -trophinames]))[1:2],
as.vector(as.matrix(g[-trophinames, -trophinames])))
temp <- temp[(temp[, 3] > 0) & !is.na(temp[, 3]), ]
temp <- temp[sort.list(temp[, 1]), ]
g_df <- data.frame(rows = temp[, 1], cols = temp[, 2], freqint = temp[,
3])
g_df$id <- 1:length(g_df[, 1])
g_df <- data.frame(id = g_df[, 4], rows = g_df[, 1], cols = g_df[,
2], freqint = g_df[, 3])
g_df_ <- melt(g_df, id = c(1, 4))
xy_s <- data.frame(value = unique(g_df_$value), x = rnorm(length(unique(g_df_$value))),
y = TrophInd(g, Import = import, Export = export)[, 1])
g_df_2 <- merge(g_df_, xy_s, by = "value")
}
# plotting
p <- ggplot(g_df_2, aes(x, y)) + geom_point(size = 5) + geom_line(aes(size = freqint,
group = id)) + geom_text(size = 3, hjust = 1.5, aes(label = value)) +
theme_bw() + opts(panel.grid.major = theme_blank(), panel.grid.minor = theme_blank(),
axis.text.x = theme_blank(), axis.text.y = theme_blank(), axis.title.x = theme_blank(),
axis.title.y = theme_blank(), axis.ticks = theme_blank(), panel.border = theme_blank(),
legend.position = "none")
p # return graph
} else if (method == "igraph") {
if (class(g) != "igraph")
stop("object must be of class 'igraph.'")
# data preparation from igraph object
g_ <- get.edgelist(g)
g_df <- as.data.frame(g_)
g_df$id <- 1:length(g_df[, 1])
g_df <- melt(g_df, id = 3)
xy_s <- data.frame(value = unique(g_df$value), x = vplace(length(unique(g_df$value))),
y = vplace(length(unique(g_df$value))))
g_df2 <- merge(g_df, xy_s, by = "value")
# plotting
p <- ggplot(g_df2, aes(x, y)) + geom_point(size = 2) + geom_line(size = 0.3,
aes(group = id, linetype = id)) + geom_text(size = 3, hjust = 1.5,
aes(label = value)) + theme_bw() + opts(panel.grid.major = theme_blank(),
panel.grid.minor = theme_blank(), axis.text.x = theme_blank(), axis.text.y = theme_blank(),
axis.title.x = theme_blank(), axis.title.y = theme_blank(), axis.ticks = theme_blank(),
panel.border = theme_blank(), legend.position = "none")
p # return graph
} else stop(paste("do not recognize method = \"", method, "\";\nmethods are \"df\" and \"igraph\"",
sep = ""))
}
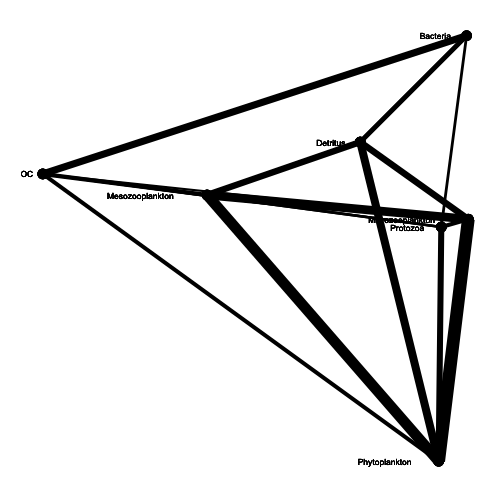
# Eg
library(NetIndices)
data(Takapoto)
gggraph(Takapoto, vplace = rnorm, method = "df", trophic = "TRUE", trophinames = c(8:10),
import = "CO2", export = c("CO2", "Sedimentation", "Grazing"))
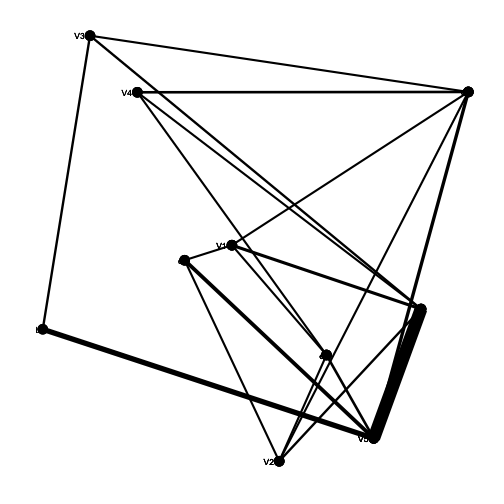
plants <- round(rlnorm(n = 5, meanlog = 2, sdlog = 1))
animals <- round(rlnorm(n = 5, meanlog = 2, sdlog = 1))
plants <- plants * (100/sum(plants))
animals <- animals * (100/sum(animals))
z <- r2dtable(1, animals, plants) # if errors, rerun again until no error
z <- as.data.frame(z[[1]])
rownames(z) <- c("a", "b", "c", "d", "e")
gggraph(z, vplace = rnorm, method = "df", trophic = "FALSE")