So, I’ve blogged about this topic before, way back on 5 Jan this year.
Matt Helmus, a postdoc in the Wootton lab at the University of Chicago, published a paper with Anthony Ives in Ecological Monographs this year (abstract here). The paper addressed a new statistical approach to phylogenetic community structure.
As I said in the original post, part of the power of the PGLMM (phylogenetic generalized linear mixed models) approach is that you don’t have to conduct quite so many separate statistical tests as with the previous null model/randomization approach.
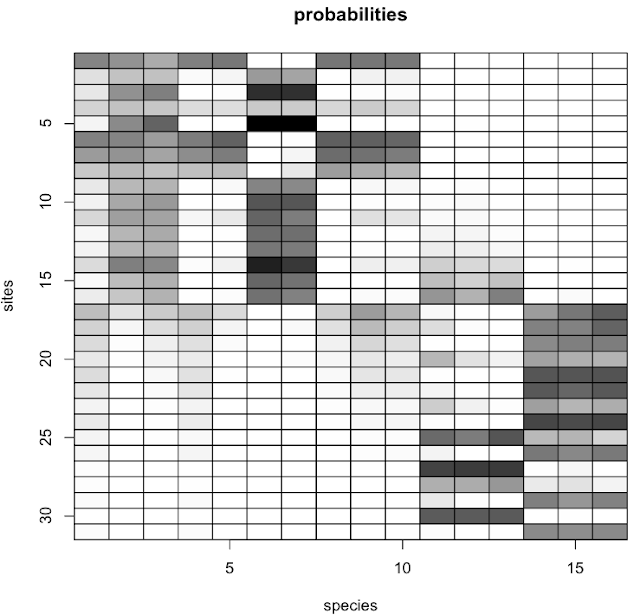
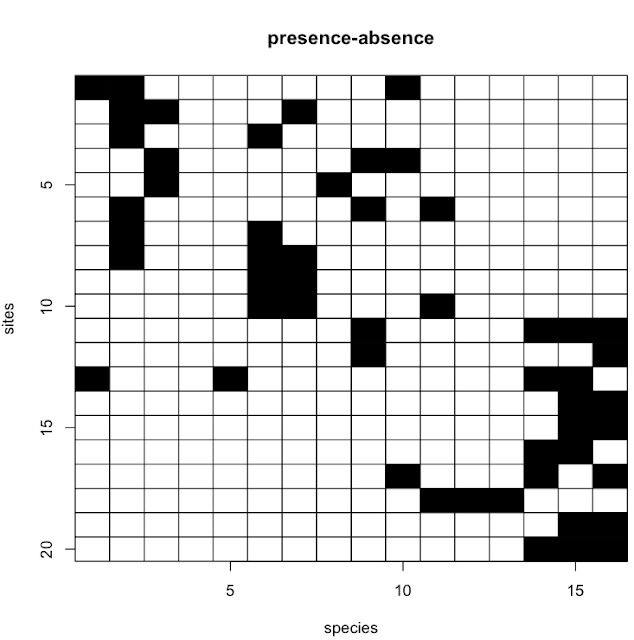
Their original code was written in Matlab. Here I provide the R code that Matt has so graciously shared with me. There are four functions and a fifth file has an example use case. The example and output are shown below.
Look for the inclusion of Matt’s PGLMM to the picante R package in the future.
Here are links to the files as GitHub gists:
PGLMM.data.R: https://gist.github.com/1278205
PGLMM.fit.R: https://gist.github.com/1284284
PGLMM.reml.R: https://gist.github.com/1284287
PGLMM.sim.R: https://gist.github.com/1284288
PGLMM_example.R: https://gist.github.com/1284442
Enjoy!
The example