UPDATE: pkg API has changed - updated the post below to work with the current CRAN version, submitted 2016-08-02
I was at a hackathon focused on Ocean Biogeographic Information System (OBIS) data back in November last year in Belgium. One project idea was to make it easier to get at data based on one or more marine regions. I was told that Marineregions.org is often used for shape files to get different regions to then do other work with.
During the event I started a package mregions. Here I’ll show how to get different marine regions, then use those outputs to get species occurrence data.
We’ll use WKT (Well-Known Text) to define spatial dimensions in this post. If you don’t know what it is, Wikipedia to the rescue: https://en.wikipedia.org/wiki/Well-known_text
Install
install.packages("mregions")
devtools::install_github("iobis/robis")
Or get the dev version
devtools::install_github("ropenscilabs/mregions")
library("mregions")
Get list of place types
res <- mr_place_types()
head(res$type)
#> [1] "Town" "Arrondissement"
#> [3] "Department" "Province (administrative)"
#> [5] "Country" "Continent"
Get Marineregions records by place type
res <- mr_records_by_type(type = "EEZ")
head(res)
#> MRGID gazetteerSource
#> 1 3293 Maritime Boundaries Geodatabase, Flanders Marine Institute
#> 2 5668 Maritime Boundaries Geodatabase, Flanders Marine Institute
#> 3 5669 Maritime Boundaries Geodatabase, Flanders Marine Institute
#> 4 5670 Maritime Boundaries Geodatabase, Flanders Marine Institute
#> 5 5672 Maritime Boundaries Geodatabase, Flanders Marine Institute
#> 6 5673 Maritime Boundaries Geodatabase, Flanders Marine Institute
#> placeType latitude longitude minLatitude minLongitude maxLatitude
#> 1 EEZ 51.46483 2.704458 51.09111 2.238118 51.87000
#> 2 EEZ 53.61508 4.190675 51.26203 2.539443 55.76500
#> 3 EEZ 54.55970 8.389231 53.24281 3.349999 55.91928
#> 4 EEZ 40.87030 19.147094 39.63863 18.461940 41.86124
#> 5 EEZ 42.94272 29.219062 41.97820 27.449580 43.74779
#> 6 EEZ 43.42847 15.650844 41.62201 13.001390 45.59079
#> maxLongitude precision preferredGazetteerName
#> 1 3.364907 58302.49 Belgian Exclusive Economic Zone
#> 2 7.208364 294046.10 Dutch Exclusive Economic Zone
#> 3 14.750000 395845.50 German Exclusive Economic Zone
#> 4 20.010030 139751.70 Albanian Exclusive Economic Zone
#> 5 31.345280 186792.50 Bulgarian Exclusive Economic Zone
#> 6 18.552360 313990.30 Croatian Exclusive Economic Zone
#> preferredGazetteerNameLang status accepted
#> 1 English standard 3293
#> 2 English standard 5668
#> 3 English standard 5669
#> 4 English standard 5670
#> 5 English standard 5672
#> 6 English standard 5673
Get and search region names
(res <- mr_names())
#> # A tibble: 676 x 4
#> name
#> <chr>
#> 1 Morocco:elevation_10m
#> 2 Emodnet:emodnet1x1grid
#> 3 Emodnet:grid
#> 4 Morocco:dam
#> 5 WoRMS:azmp_sections
#> 6 Morocco:accomodationcapacity
#> 7 Morocco:admin_boundary
#> 8 Lifewatch:ovam_afvalverwerking
#> 9 Eurobis:eurobis_points
#> 10 Morocco:roads
#> # ... with 666 more rows, and 3 more variables: title <chr>,
#> # name_first <chr>, name_second <chr>
mr_names_search(res, q = "IHO")
#> # A tibble: 5 x 4
#> name
#> <chr>
#> 1 MarineRegions:iho
#> 2 MarineRegions:iho_quadrants_20150810
#> 3 World:iosregions
#> 4 MarineRegions:eez_iho_union_v2
#> 5 Belgium:vl_venivon
#> # ... with 3 more variables: title <chr>, name_first <chr>,
#> # name_second <chr>
Get a region - geojson
res <- mr_geojson(name = "Turkmen Exclusive Economic Zone")
class(res)
#> [1] "mr_geojson"
names(res)
#> [1] "type" "totalFeatures" "features" "crs"
#> [5] "bbox"
Get a region - shp
res <- mr_shp(name = "Belgian Exclusive Economic Zone")
class(res)
#> [1] "SpatialPolygonsDataFrame"
#> attr(,"package")
#> [1] "sp"
Get OBIS EEZ ID
res <- mr_names()
res <- res[grepl("eez", res$name, ignore.case = TRUE),]
mr_obis_eez_id(res$title[2])
#> [1] 84
Convert to WKT
From geojson or shp. Here, geojson
res <- mr_geojson(key = "MarineRegions:eez_33176")
mr_as_wkt(res, fmt = 5)
#> [1] "MULTIPOLYGON (((41.573732 -1.659444, 45.891882 ... cutoff
Get regions, then OBIS data
Using Well-Known Text
Both shp and geojson data returned from region_shp() and region_geojson(), respectively, can be passed to as_wkt() to get WKT.
shp <- mr_shp(name = "Belgian Exclusive Economic Zone")
wkt <- mr_as_wkt(shp)
library('httr')
library('data.table')
args <- list(scientificname = "Abra alba", geometry = wkt, limit = 100)
res <- httr::GET('https://api.iobis.org/occurrence', query = args)
xx <- data.table::setDF(data.table::rbindlist(httr::content(res)$results, use.names = TRUE, fill = TRUE))
xx <- xx[, c('scientificName', 'decimalLongitude', 'decimalLatitude')]
names(xx)[2:3] <- c('longitude', 'latitude')
Plot
library('leaflet')
leaflet() %>%
addTiles() %>%
addCircleMarkers(data = xx) %>%
addPolygons(data = shp)
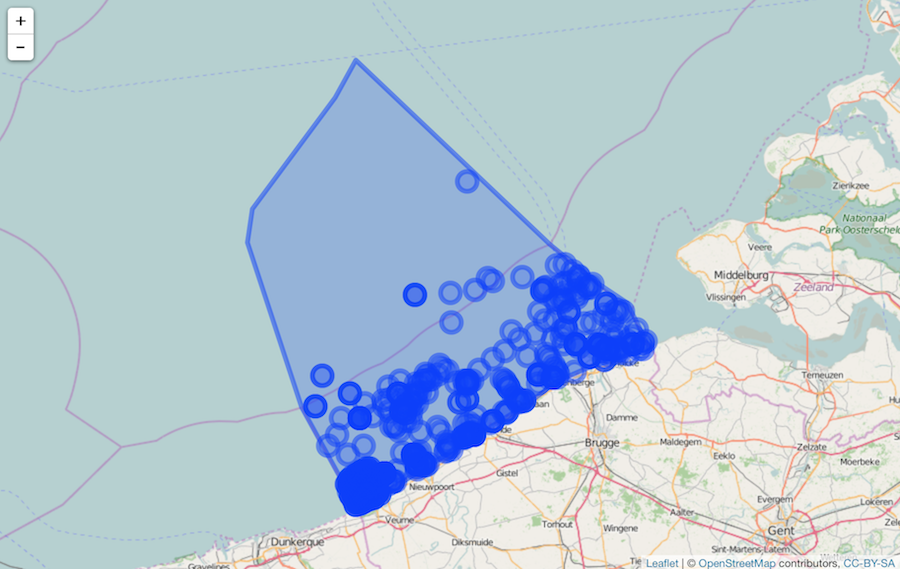
Using EEZ ID
EEZ is a Exclusive Economic Zone
(eez <- mr_obis_eez_id("Belgian Exclusive Economic Zone"))
#> [1] 59
You currently can’t search for OBIS occurrences by EEZ ID, but hopefully soon…
Dealing with bigger WKT
What if you’re WKT string is super long? It’s often a problem because some online species occurrence databases that accept WKT to search by geometry bork due to limitations on length of URLs if your WKT string is too long (about 8000 characters, including remainder of URL). One way to deal with it is to reduce detail - simplify.
install.packages("rmapshaper")
Using rmapshaper we can simplify a spatial object, then search with that.
shp <- mr_shp(name = "Dutch Exclusive Economic Zone")
Visualize
leaflet() %>%
addTiles() %>%
addPolygons(data = shp)
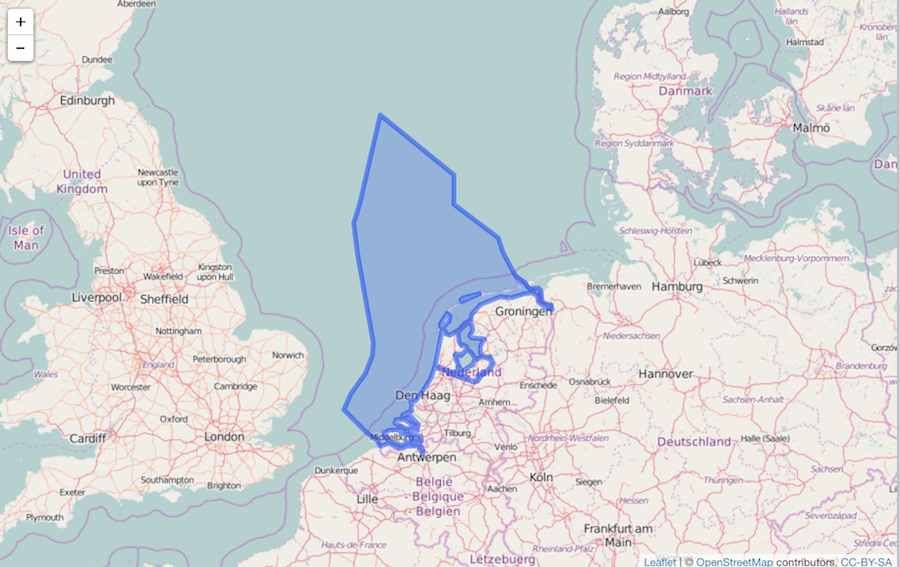
Simplify
library("rmapshaper")
shp <- ms_simplify(shp)
It’s simplified:
leaflet() %>%
addTiles() %>%
addPolygons(data = shp)
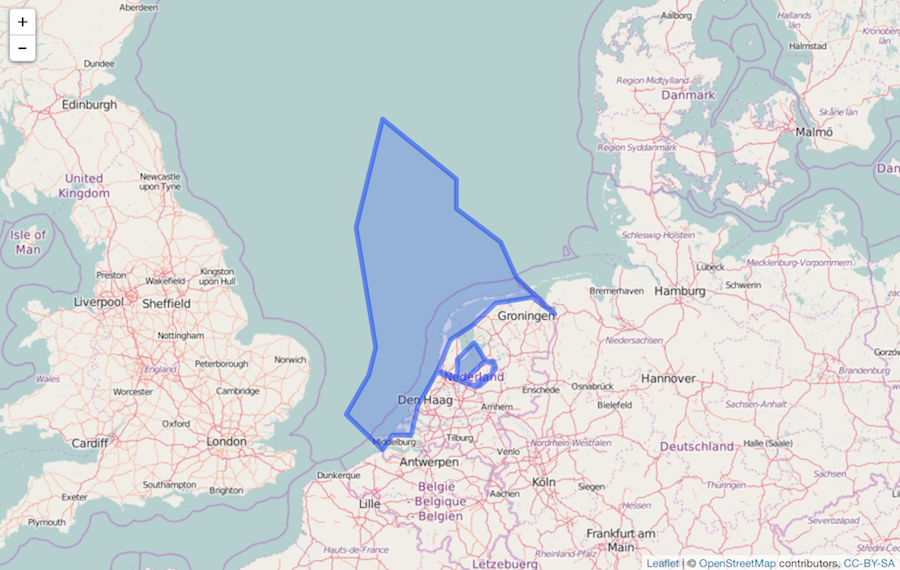
Pass to GBIF
if (!requireNamespace("rgbif")) {
install.packages("rgbif")
}
library("rgbif")
occ_search(geometry = mr_as_wkt(shp), limit = 400)
#> Records found [2395936]
#> Records returned [400]
#> No. unique hierarchies [17]
#> No. media records [3]
#> Args [geometry=POLYGON ((7.2083632399999997 53.2428091399999985,
#> 6.6974995100000001 53.4619365499999972, 5.89083650, limit=400,
#> offset=0, fields=all]
#> # A tibble: 400 x 102
#> name key decimalLatitude decimalLongitude
#> <chr> <int> <dbl> <dbl>
#> 1 Haematopus ostralegus 1265900666 52.13467 4.21306
#> 2 Limosa limosa 1265577408 53.03249 4.88665
#> 3 Corvus splendens 1269536058 51.98297 4.12982
#> 4 Corvus splendens 1269536065 51.98297 4.12982
#> 5 Lanius excubitor 1211320606 52.57223 4.62569
#> 6 Lanius excubitor 1211320862 52.67419 4.63645
#> 7 Lanius excubitor 1211320806 53.05790 4.72974
#> 8 Lanius excubitor 1211321040 52.57540 4.63651
#> 9 Lanius excubitor 1211320590 52.41180 5.19500
#> 10 Lanius excubitor 1211320401 52.57535 4.63654
#> # ... with 390 more rows, and 98 more variables: issues <chr>,
#> # datasetKey <chr>, publishingOrgKey <chr>, publishingCountry <chr>,
#> # protocol <chr>, lastCrawled <chr>, lastParsed <chr>, extensions <chr>,
#> # basisOfRecord <chr>, taxonKey <int>, kingdomKey <int>,
#> # phylumKey <int>, classKey <int>, orderKey <int>, familyKey <int>,
#> # genusKey <int>, speciesKey <int>, scientificName <chr>, kingdom <chr>,
#> # phylum <chr>, order <chr>, family <chr>, genus <chr>, species <chr>,
#> # genericName <chr>, specificEpithet <chr>, taxonRank <chr>,
#> # dateIdentified <chr>, coordinateUncertaintyInMeters <dbl>, year <int>,
#> # month <int>, day <int>, eventDate <chr>, modified <chr>,
#> # lastInterpreted <chr>, references <chr>, identifiers <chr>,
#> # facts <chr>, relations <chr>, geodeticDatum <chr>, class <chr>,
#> # countryCode <chr>, country <chr>, rightsHolder <chr>,
#> # identifier <chr>, informationWithheld <chr>, verbatimEventDate <chr>,
#> # datasetName <chr>, gbifID <chr>, collectionCode <chr>,
#> # verbatimLocality <chr>, occurrenceID <chr>, taxonID <chr>,
#> # license <chr>, recordedBy <chr>, catalogNumber <chr>,
#> # http...unknown.org.occurrenceDetails <chr>, institutionCode <chr>,
#> # rights <chr>, eventTime <chr>, identificationID <chr>,
#> # individualCount <int>, continent <chr>, stateProvince <chr>,
#> # nomenclaturalCode <chr>, locality <chr>, language <chr>,
#> # taxonomicStatus <chr>, type <chr>, preparations <chr>,
#> # disposition <chr>, originalNameUsage <chr>,
#> # bibliographicCitation <chr>, identifiedBy <chr>, sex <chr>,
#> # lifeStage <chr>, otherCatalogNumbers <chr>, habitat <chr>,
#> # georeferencedBy <chr>, vernacularName <chr>, elevation <dbl>,
#> # minimumDistanceAboveSurfaceInMeters <chr>, dynamicProperties <chr>,
#> # samplingEffort <chr>, organismName <chr>, georeferencedDate <chr>,
#> # georeferenceProtocol <chr>, georeferenceVerificationStatus <chr>,
#> # organismID <chr>, ownerInstitutionCode <chr>, samplingProtocol <chr>,
#> # datasetID <chr>, accessRights <chr>, georeferenceSources <chr>,
#> # elevationAccuracy <dbl>, depth <dbl>, depthAccuracy <dbl>,
#> # waterBody <chr>