Functions can have no parameters, or have a lot of parameters, or somewhere in between. How many parameters is too many? Does it even matter how many parameters there are in a function?
There’s AFAIK no “correct” answer to this question. And surely the “best practice” varies among programming languages. What do folks say about this and what should we be doing in R?
From other languages
Many of the blog posts and SO posts on this topic cite the book Clean Code by “Uncle Bob”. I’ve not read the book, but it sounds worth a read.
Some of the arguments go like: too many arguments can …
- makes it easier to pass arguments in the wrong order
- reduce code readability
- make it harder to test a function; it’s difficult/time consuming to test all various combinations of arguments work together
An analysis was done in 2018 of php open source projects, and they found that the most common number of parameters was 5; functions with 10 parameters or more were found in <20% of projects.
On the other side, some argue that you shouldn’t worry so much about the correct number of parameters, but rather make sure that all the parameters make sense, and are documented and tested.
To the extreme, a number of people quote the Clean Code book:
The ideal number of arguments for a function is zero
Some general threads on this topic:
Data
Data for this post, created below, is in the github repo sckott/howmanyparams.
What about R?
What do the data show in the R language? Just like the blog post on php above, let’s have a look at a lot of R packages to get a general data informed consensus on how many parameters are used per function.
It’s incredibly likely that there is a better way to do what I’ve done below; but this is my hacky way of getting to the data.
What I’ve done in words:
- Get a list of all available package names on CRAN
- Install about half of them (didn’t do all cause it takes time and I don’t think I need all 15K packages to get a good answer)
- List the exported functions in each package
- Count the arguments (parameters) per function in each package
- Visualize the results
I ended up using 82489 functions across 4777 packages
Load packages
library(plyr)
library(dplyr)
library(tibble)
library(ggplot2)
Use a different path from my actual R library location to not pollute my current setup, and put binaries into a temporary directory so they are cleaned up on exiting R.
path <- "/some/path"
binaries <- file.path(tempdir(), "binaries")
dir.create(path)
dir.create(binaries)
.libPaths(path)
.libPaths() # check that the path was set
Function do_one() to run on each package:
- try to load the package
- if not found install it
- get a vector of the exported functions in the package
- count how many arguments each function has, make a data.frame
- unload the package namespace
do_one <- function(pkg) {
if (!requireNamespace(pkg))
install.packages(pkg, quiet=TRUE, verbose=FALSE, destdir = binaries)
on.exit(unloadNamespace(pkg))
funs <- paste0(pkg, "::", getNamespaceExports(pkg))
enframe(vapply(funs, function(w) {
tt <- tryCatch(parse(text = w), error = function(e) e)
if (!inherits(tt, "error")) length(suppressWarnings(formals(eval(tt)))) else 0
}, numeric(1)))
}
do_one_safe <- failwith(tibble(), do_one)
Get a list of packages. First time running through I used available.packages() which
gets you all available packages. After installing packages though, I used
installed.packages() to get the list of packages I already installed.
# pkg_names <- unname(available.packages()[,"Package"])
pkg_names <- unname(installed.packages()[,"Package"])
Run each package through the do_one() function. This had to be stopped and
re-started a few times. This failed for quite a few packages - I wasn’t trying to get every single package, just a large set of packages to get an idea of what packages do on average.
tbls <- stats::setNames(lapply(pkg_names, do_one_safe), pkg_names)
Combine list of data.frame’s into one data.frame
df <- dplyr::bind_rows(tbls, .id = "pkg")
readr::write_csv(df, "params_per_fxn.csv")
note: you can get this data at sckott/howmanyparams
df <- readr::read_csv("~/params_per_fxn.csv")
Visualize
All functions across all packages
ggplot(df, aes(x = value)) +
geom_histogram(bins = 30) +
scale_x_continuous(limits = c(0, 30)) +
theme_grey(base_size = 15)
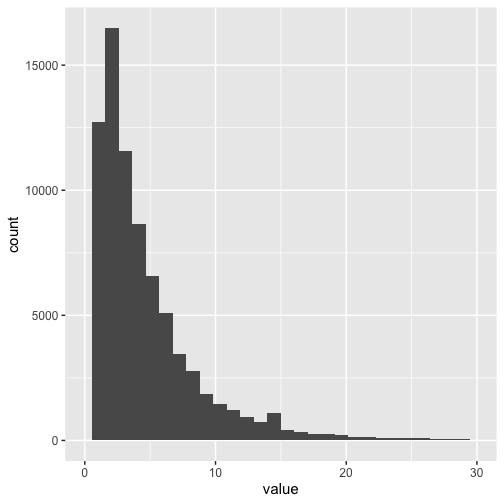
The mean number of arguments per function across all packages was 4.4, and the most common value was 3. The maximum number of arguments was 209, and there were 5306 functions (or 6.4%) with zero parameters.
Mean params across functions for each pkg
df_means <- group_by(df, pkg) %>%
summarise(mean_params = mean(value, na.rm=TRUE)) %>%
ungroup()
# arrange(df_means, desc(mean_params))
ggplot(df_means, aes(x = mean_params)) +
geom_histogram(bins = 50) +
scale_x_continuous(limits = c(0, 30)) +
theme_grey(base_size = 15)
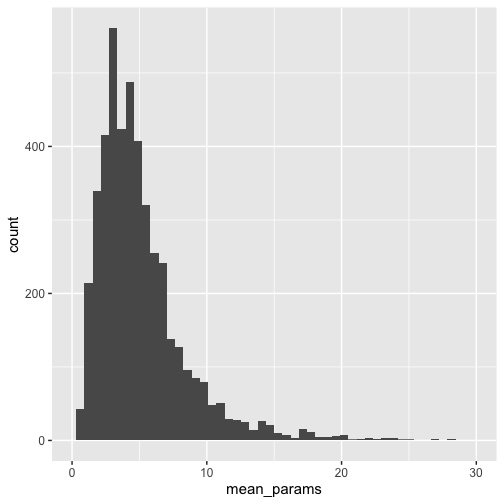
Taking the mean within each package first pulls the number of arguments to the right some, with a mean of 5 arguments, and the most common value at 4.
Thoughts
In terms of getting around the too many arguments thing, there’s talk of using global variables, which seems like generally a bad idea; unless perhaps they are environment variables that are meant to be set by the user in non-interactive environments, etc.
Other solutions are to use ... in R, or similarly **kwargs or *args in Python (ref.), or
the newly added ... in Ruby (ref). With this approach you could have very few parameters
defined in the function, and then internally within the function handle any parameter
filtering, etc. The downside of this in R is that you don’t get the automated
checks making sure all function arguments are documented, and there’s no documented
arguments that don’t exist in the function.
I’m not suggesting a solution is needed though; there’s probably no right answer, but rather lots of opinions.
Having said that, the average R function does use about 4 arguments, so if you keep your functions to around 4 arguments you’ll be approaching the sort of consensus of a large number of R developers.
Last, I should admit that some of the functions in my packages have quite a lot
of parameters - which was sort of the motivation for this post - that is, to explore
what most functions do. For example, brranching::phylomatic has 13 parameters,
three functions in the crevents package have 24 parameters … and I wonder
about these types of functions. Should I refactor? Or is it good enough to make
sure these functions are thoroughly documented and tested?